1-s2.0-S0006291X20320593-gr3 lrg
Description:
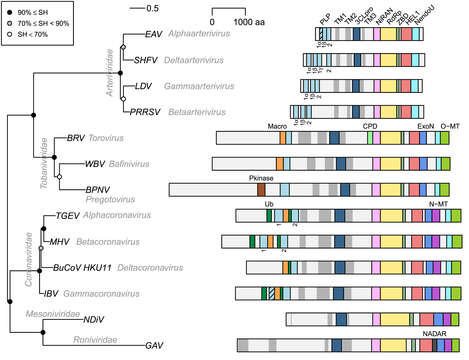
All images in this article were uploaded in the JPEG format even though it consists of non-photographic data. This information could be stored more efficiently or accurately in the PNG or SVG format. If possible, please upload a PNG or SVG version of this image without compression artifacts, derived from a non-JPEG source (or with existing artifacts removed). After doing so, please tag the JPEG version with.mw-parser-output.nowrap,.mw-parser-output.nowrap a:before,.mw-parser-output.nowrap.selflink:before{white-space:nowrap}{{Superseded|NewImage.ext}} and remove this tag. This tag should not be applied to photographs or scans. For more information, see {{BadJPEG}}. Description: English: Phylogeny and pp1ab domain organization of selected nidoviruses. Representatives of 13 lineages of nidoviruses that infect vertebrates (three virus families) and invertebrates (two virus families) are depicted. Names of taxa, families and genera, are indicated in grey italic font, and names of viruses are given as acronyms: EAV, equine arteritis virus; SHFV, simian hemorrhagic fever virus; LDV, lactate dehydrogenase-elevating virus; PRRSV, porcine reproduction respiratory syndrome virus; BRV, Breda virus; WBV, white bream virus; "BPNV, ball python nidovirus;" TGEV, porcine transmissible gastroenteritis virus; MHV, mouse hepatitis virus; BuCoV_HKU11, bulbul coronavirus HKU11; IBV, avian infectious bronchitis virus; NDiV, Nam Dinh virus; GAV, gill-associated virus. Midpoint rooted phylogeny was reconstructed based on Viralis multiple sequence alignment [92] of the conserved core of RdRp, using IQ-Tree 1.5.5 [93] with automatically selected the rtREV + F + I + G4 evolutionary model. To estimate branch support, SH-like approximate likelihood ratio test with 1000 replicates was conducted. Polyproteins pp1ab are shown as light grey bars; they are autoproteolytically processed to nsps that were identified only for few nidoviruses and omitted here (see also Fig. 1). TM domains are shown as dark grey bars; TM helices were predicted by TMHMM2.0c [94] and clustered if separated by less than 300 aa (less than 180 aa for arteri- and tobaniviruses). Other selected domains, whose coordinates were obtained from the Viralis database [92], are shown as colored bars; proteolytically inactive PLP domains are indicated by stripes on bars; indices of PLP domains are specified below the bars. "Pkinase, protein kinase [25]; CPD, cyclic phosphodiestarase known also as 2′,5′-phosphodiesterase, 2′PDE [58,95]; NADAR, domain involved in the utilization of NAD and ADP-ribose derivatives [96]; for other domains, see Fig. 1 in the source. Date: 29 January 2021. Source: Anastasia A. Gulyaeva, Alexander E. Gorbalenya, A nidovirus perspective on SARS-CoV-2, Biochemical and Biophysical Research Communications, Volume 538, 2021, Pages 24-34, ISSN 0006-291X, https://doi.org/10.1016/j.bbrc.2020.11.015. Author: Anastasia A.Gulyaeva and Alexander E.Gorbalenya.
Included On The Following Pages:
This image is not featured in any collections.
Source Information
- license
- cc-by-3.0
- copyright
- Anastasia A.Gulyaeva and Alexander E.Gorbalenya
- creator
- Anastasia A.Gulyaeva and Alexander E.Gorbalenya
- source
- Anastasia A. Gulyaeva, Alexander E. Gorbalenya, A nidovirus perspective on SARS-CoV-2, Biochemical and Biophysical Research Communications, Volume 538, 2021, Pages 24-34, ISSN 0006-291X,
- original
- original media file
- visit source
- partner site
- Wikimedia Commons
- ID