Journal.ppat.1000772.g001
Description:
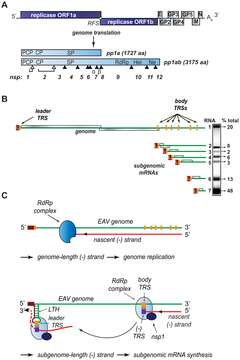
Description: English: Organization and expression of the polycistronic arterivirus equine arteritis virus (EAV) +RNA genome. (A) Top: EAV genome organization, showing the 5′-proximal replicase open reading frames (ORFs), as well as the downstream ORFs encoding the viral structural proteins envelope (E), membrane (M), nucleocapsid (N), and glycoproteins (GP) 2–5 and the 3′ poly(A) tail (An). Bottom: overview of the pp1a and pp1ab replicase polyproteins that result from genome translation, which requires an ORF1a/1b ribosomal frameshift (RFS) to produce pp1ab. Arrowheads represent sites cleaved by the three virus-encoded proteases (open for autoproteolytically processed ones, closed for sites processed by the main proteinase in nsp4). The resulting nonstructural proteins (nsp) are numbered. The key viral enzymatic domains such as the nsp1 papain-like cysteine proteinase β (PCP), nsp2 cysteine proteinase (CP), nsp4 serine proteinase (SP), nsp9 viral RNA-dependent RNA polymerase (RdRp), nsp10 helicase (Hel), and nsp11 endoribonuclease (Ne) are indicated. (B) Overview of viral mRNA species produced in EAV-infected cells. The ORFs expressed from the respective mRNAs are shown in gray, and the 5′ leader sequence is depicted in dark red. The orange boxes indicate the positions of transcription-regulating sequences (TRS). The gel hybridization image on the right is representative of the wild-type accumulation levels of the seven EAV mRNAs at the time point used for analysis in the study (see text for details). The amount of each mRNA, determined by quantitative phosphorimager analysis, is indicated as percentage of the total amount of viral mRNA. (C) Model for EAV replication and transcription. Continuous minus-strand RNA synthesis yields a genome-length minus strand template for genome replication, a process for which nsp1 is dispensable. Discontinuous minus-strand RNA synthesis results in a nested set of subgenome-length minus strands that serve as templates for sg mRNA synthesis (see text for details). Nsp1 is crucial for this process, which is also guided by a base pairing interaction between the TRS complement [(−)TRS] at the 3′ end of the nascent minus-strand and the genomic leader TRS, present in a RNA hairpin structure (LTH). Date: 19 February 2010. Source: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1000772. Author: Danny D. Nedialkova, Alexander E. Gorbalenya, Eric J. Snijder.
Included On The Following Pages:
This image is not featured in any collections.
Source Information
- license
- cc-by-3.0
- copyright
- Danny D. Nedialkova, Alexander E. Gorbalenya, Eric J. Snijder
- creator
- Danny D. Nedialkova, Alexander E. Gorbalenya, Eric J. Snijder
- source
- https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1000772
- original
- original media file
- visit source
- partner site
- Wikimedia Commons
- ID