en
names in breadcrumbs

 類型
類型 正黏液病毒科(Orthomyxoviridae,希臘文Orthos有「正確,直」之意;myxo有「粘液」之意[1])屬於負鏈RNA病毒,共有六個屬,此科的病毒可感染脊椎動物。造成流行性感冒的病毒正是正黏液病毒科的一員,屬於負鏈RNA病毒。正黏液病毒科可分為四屬病毒,三屬為流行性感冒病毒,分為A型、B型及C型流行性感冒病毒,以及传染性鲑鱼贫血病毒屬和托高土病毒屬。流行性感冒病毒可以感染人、馬、豬及禽類(參見禽流感),其中A型流行性感冒病毒可以感染人、豬、馬及鳥類,而B型流行性感冒,僅可感染人類。C型流行性感冒病毒僅感染人、豬,造成之病害較少。 A型及B型流行性感冒病毒之感染力非常強,傳播速度非常快,可以稱之為最重要的疾病之一,感染範圍遍及世界各大洲。水禽類,尤其是候鳥,為各不同血清型病毒之帶原者及病毒重組之動物。传染性鲑鱼贫血病毒則鮭魚,托高土病毒則可以感染脊椎動物及非脊椎動物,如蚊子和海蝨[2][3][4]。
本科中有三屬為流感病毒,其分類方法依照其表面NP及M抗原相異性:
按系統分類學,此科屬於RNA病毒的ssRNA(-),其關聯物種及血清型如下表所示:
正黏液病毒屬別、種及血清型 屬 種(*為模式種) 血清型或亞型 寄主 A型流感病毒 A型流感病毒* H1N1、H1N2、H2N2、H3N1、H3N2、H3N8流行性感冒病毒係依其基質蛋白(matrix protein)及內部核糖核蛋白(ribonucleoprotein;可溶性抗原)之抗原特性而分為A、B及C三種,外層蛋白包括血球凝集素(hemagglutinin,HA)及神經氨酸酶(neuraminidase, NA)兩種,也為組成流行性感冒的抗原成份。流行性感冒病毒依其血球凝集素及神經胺酸酶之抗原性不同可再分為數種亞型,現今已知血球凝集素已有十六種(H1-H16),神經胺酸酶則有九種。C型流行性感冒之化學性及結構性與 A 及 B 型流行性感冒皆十分相似,但它缺少神經胺酸酶,只有血球凝集素酶一種,在病毒表面上具有一特殊的接受器(receptor),使病毒表面呈蜂窩狀,而 A 型及 B 型流行性感冒很少呈蜂窩狀。
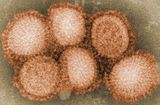
流行性感冒病毒在電子顯微鏡下有絲狀及多形性二種,但大部份為多形性,負染色時常為 70 - 120 nm 之圓形顆粒。新分離的流行性感冒病毒顆粒,其大小及形狀皆不一致,絲狀者經常可長達幾百奈米(nanometer),甚至長達 4,000 nm,有時可以見到一些奇形怪狀者,流行性感冒病毒與其它病毒不同的是它的整個病毒表面有約略等距間隔的突起,一種為長約 10 - 14 nm,寬約 4 nm ,橫切面為三角形,含有血球凝集素的桿狀突起,另一為長約 9 nm,寬約 5 nm,含有神經胺酸酶的圓柱狀突起。
在針狀突起層下面為一包圍基質蛋白及核酸蛋白之脂質被膜, A型流行性感冒病毒之核蛋白衣(nucleocapsid)含有與病毒 RNA 相連的 RNA 聚合酶, B 型及 C 型流行性感冒病毒之核蛋白衣亦可能有相同的結構。病毒封套(envelope)厚度為 6 至 8 nm,當封套破裂或染色液滲入時,可能會看到核酸呈摺疊狀平行的條狀結構。以清潔劑處理也可能會得到大小不等的極度卷曲形狀的核蛋白衣。
C 型流行性感冒病毒外型與 A或B型結構大致相同,不容易區分,有時可見病毒表面有六角型的結構,通常在電子顯微鏡下,同一視野中可能會同時見到這兩種不同形狀的病毒顆粒。
血球凝集素可附著於不同動物的紅血球,而使紅血球凝集,而神經胺酸酶與病毒離開寄主細胞有關,神經胺酸酶會分解膜上的神經胺酸,而使病毒由寄主細胞脫離感染其他細胞。 C 型流行性感冒病毒也含有可破壞流行性感冒接受器的酶,但它與神經胺酸酶不同。
Viruses of this family contain 6 to 8 segments of linear negative-sense single stranded RNA.[7]
The total genome length is 12000–15000 nucleotides (nt). The largest segment 2300–2500 nt; of second largest 2300–2500 nt; of third 2200–2300 nt; of fourth 1700–1800 nt; of fifth 1500–1600 nt; of sixth 1400–1500 nt; of seventh 1000–1100 nt; of eighth 800–900 nt. Genome sequence has terminal repeated sequences; repeated at both ends. Terminal repeats at the 5'-end 12–13 nucleotides long. Nucleotide sequences of 3'-terminus identical; the same in genera of same family; most on RNA (segments), or on all RNA species. Terminal repeats at the 3'-end 9–11 nucleotides long. Encapsidated nucleic acid is solely genomic. Each virion may contain defective interfering copies.
The following applies for Influenza A viruses, although other influenza strains are very similar in structure:[8]
The influenza A virus particle or virion is 80–120 nm in diameter and usually roughly spherical, although filamentous forms can occur.[9] Unusually for a virus, the influenza A genome is not a single piece of nucleic acid; instead, it contains eight pieces of segmented negative-sense RNA (13.5 kilobases total), which encode 11 proteins (HA, NA, NP, M1, M2, NS1, NEP, PA, PB1, PB1-F2, PB2).[10] The best-characterised of these viral proteins are hemagglutinin and neuraminidase, two large glycoproteins found on the outside of the viral particles. Neuraminidase is an enzyme involved in the release of progeny virus from infected cells, by cleaving sugars that bind the mature viral particles. By contrast, hemagglutinin is a lectin that mediates binding of the virus to target cells and entry of the viral genome into the target cell.[11] The hemagglutinin (H) and neuraminidase (N) proteins are targets for antiviral drugs.[12] These proteins are also recognised by antibodies, i.e. they are antigens.[13] The responses of antibodies to these proteins are used to classify the different serotypes of influenza A viruses, hence the H and N in H5N1.
Typically, influenza is transmitted from infected mammals through the air by coughs or sneezes, creating aerosols containing the virus, and from infected birds through their droppings. Influenza can also be transmitted by saliva, nasal secretions, feces and blood. Infections occur through contact with these bodily fluids or with contaminated surfaces. Flu viruses can remain infectious for about one week at human body temperature, over 30 days at 0 °C(32 °F), and indefinitely at very low temperatures (such as lakes in northeast Siberia). They can be inactivated easily by disinfectants and detergents.[14][15][16]
The viruses bind to a cell through interactions between its hemagglutinin glycoprotein and sialic acid sugars on the surfaces of epithelial cells in the lung and throat (Stage 1 in infection figure).[17] The cell imports the virus by endocytosis. In the acidic endosome, part of the haemagglutinin protein fuses the viral envelope with the vacuole's membrane, releasing the viral RNA (vRNA) molecules, accessory proteins and RNA-dependent RNA polymerase into the cytoplasm (Stage 2).[18] These proteins and vRNA form a complex that is transported into the cell nucleus, where the RNA-dependent RNA polymerase begins transcribing complementary positive-sense cRNA (Steps 3a and b).[19] The cRNA is either exported into the cytoplasm and translated (step 4), or remains in the nucleus. Newly synthesised viral proteins are either secreted through the Golgi apparatus onto the cell surface (in the case of neuraminidase and hemagglutinin, step 5b) or transported back into the nucleus to bind vRNA and form new viral genome particles (step 5a). Other viral proteins have multiple actions in the host cell, including degrading cellular mRNA and using the released nucleotides for vRNA synthesis and also inhibiting translation of host-cell mRNAs.[20]
Negative-sense vRNAs that form the genomes of future viruses, RNA-dependent RNA transcriptase, and other viral proteins are assembled into a virion. Hemagglutinin and neuraminidase molecules cluster into a bulge in the cell membrane. The vRNA and viral core proteins leave the nucleus and enter this membrane protrusion (step 6). The mature virus buds off from the cell in a sphere of host phospholipid membrane, acquiring hemagglutinin and neuraminidase with this membrane coat (step 7).[21] As before, the viruses adhere to the cell through hemagglutinin; the mature viruses detach once their neuraminidase has cleaved sialic acid residues from the host cell.[17] After the release of new influenza virus, the host cell dies.
Orthomyxoviridae viruses are one of the only RNA viruses that replicate in the nucleus. This is because the machinery of orthomyxo viruses cannot make their own mRNAs. They use cellular RNAs as primers for initiating the viral mRNA synthesis in a process known as cap-snatching.[22] Once in the nucleus, the RNA Polymerase Protein PB2 finds a cellular pre-mRNA and binds to its 5' capped end. Then RNA Polymerase PA cleaves off the cellular mRNA near the 5' end and uses this capped fragment as a primer for transcribing the rest of the viral RNA genome in viral mRNA.[23] This is due to the need of mRNA to have a 5' cap in order to be recognized by the cell's ribosome for translation.
Since RNA proofreading enzymes are absent, the RNA-dependent RNA transcriptase makes a single nucleotide insertion error roughly every 10 thousand nucleotides, which is the approximate length of the influenza vRNA. Hence, nearly every newly manufactured influenza virus will contain a mutation in its genome.[24] The separation of the genome into eight separate segments of vRNA allows mixing (reassortment) of the genes if more than one variety of influenza virus has infected the same cell (superinfection). The resulting alteration in the genome segments packaged into viral progeny confers new behavior, sometimes the ability to infect new host species or to overcome protective immunity of host populations to its old genome (in which case it is called an antigenic shift).[13]
There are three genera of influenza virus: Influenzavirus A, Influenzavirus B and Influenzavirus C. Each genus includes only one species, or type: Influenza A virus, Influenza B virus, and Influenza C virus, respectively. Influenza A and C infect multiple species, while influenza B almost exclusively infects humans.[25][26]
Influenza A viruses are further classified, based on the viral surface proteins hemagglutinin (HA or H) and neuraminidase (NA or N). Sixteen H subtypes (or serotypes) and nine N subtypes of influenza A virus have been identified.
Further variation exists; thus, specific influenza strain isolates are identified by a standard nomenclature specifying virus type, geographical location where first isolated, sequential number of isolation, year of isolation, and HA and NA subtype.[27][28]
Examples of the nomenclature are:
The type A viruses are the most virulent human pathogens among the three influenza types and cause the most severe disease. The serotypes that have been confirmed in humans, ordered by the number of known human pandemic deaths, are:
Influenza B virus is almost exclusively a human pathogen, and is less common than influenza A. The only other animal known to be susceptible to influenza B infection is the seal.[37] This type of influenza mutates at a rate 2–3 times lower than type A[38] and consequently is less genetically diverse, with only one influenza B serotype.[25] As a result of this lack of antigenic diversity, a degree of immunity to influenza B is usually acquired at an early age. However, influenza B mutates enough that lasting immunity is not possible.[39] This reduced rate of antigenic change, combined with its limited host range (inhibiting cross species antigenic shift), ensures that pandemics of influenza B do not occur.[40]
The influenza C virus infects humans and pigs, and can cause severe illness and local epidemics.[41] However, influenza C is less common than the other types and usually seems to cause mild disease in children.[42][43]
Mammalian influenza viruses tend to be labile, but can survive several hours in mucus.[44] Avian influenza virus can survive for 100 days in distilled water at room temperature, and 200 days at 17 °C(63 °F). The avian virus is inactivated more quickly in manure, but can survive for up to 2 weeks in feces on cages. Avian influenza viruses can survive indefinitely when frozen.[44] Influenza viruses are susceptible to bleach, 70% ethanol, aldehydes, oxidizing agents, and quaternary ammonium compounds. They are inactivated by heat of 133 °F(56 °C) for minimum of 60 minutes, as well as by low pH <2.[44]
Vaccines and drugs are available for the prophylaxis and treatment of influenza virus infections. Vaccines are composed of either inactivated or live attenuated virions of the H1N1 and H3N2 human influenza A viruses, as well as those of influenza B viruses. Because the antigenicities of the wild viruses evolve, vaccines are reformulated annually by updating the seed strains. However, when the antigenicities of the seed strains and wild viruses do not match, vaccines fail to protect the vaccinees. In addition, even when they do match, escape mutants are often generated. Drugs available for the treatment of influenza include Amantadine and Rimantadine, which inhibit the uncoating of virions by interfering with M2, and Oseltamivir (marketed under the brand name Tamiflu), Zanamivir, and Peramivir, which inhibit the release of virions from infected cells by interfering with NA. However, escape mutants are often generated for the former drug and less frequently for the latter drug.[45]
台灣於1972年時首次發現家禽流行性感冒病毒感染症,血清型為H6N1 ,現今在台灣H血清型共十型 (H1,2,3,4,6,7,8,10,11,14), N血清型共八型 (N1,2,3,4,6,7,8,9) ,其組合共十五種。A型流行性感冒病毒對豬而言,可造成嚴重的增殖性壞死性肺炎。
|author-separator= (帮助) 正黏液病毒科(Orthomyxoviridae,希臘文Orthos有「正確,直」之意;myxo有「粘液」之意)屬於負鏈RNA病毒,共有六個屬,此科的病毒可感染脊椎動物。造成流行性感冒的病毒正是正黏液病毒科的一員,屬於負鏈RNA病毒。正黏液病毒科可分為四屬病毒,三屬為流行性感冒病毒,分為A型、B型及C型流行性感冒病毒,以及传染性鲑鱼贫血病毒屬和托高土病毒屬。流行性感冒病毒可以感染人、馬、豬及禽類(參見禽流感),其中A型流行性感冒病毒可以感染人、豬、馬及鳥類,而B型流行性感冒,僅可感染人類。C型流行性感冒病毒僅感染人、豬,造成之病害較少。 A型及B型流行性感冒病毒之感染力非常強,傳播速度非常快,可以稱之為最重要的疾病之一,感染範圍遍及世界各大洲。水禽類,尤其是候鳥,為各不同血清型病毒之帶原者及病毒重組之動物。传染性鲑鱼贫血病毒則鮭魚,托高土病毒則可以感染脊椎動物及非脊椎動物,如蚊子和海蝨。
本科中有三屬為流感病毒,其分類方法依照其表面NP及M抗原相異性:
甲型流感病毒:感染人、鳥,和其他爬蟲類,是大流行的元兇。 乙型流感病毒:人、海豹 丙型流感病毒:感染人、豬、狗